Decorate LFQData with Methods for transforming Intensities
Source:R/LFQDataTransformer.R
LFQDataTransformer.RdDecorate LFQData with Methods for transforming Intensities
Decorate LFQData with Methods for transforming Intensities
Methods
Method log2()
log2 transform data
Method robscale()
robust scale data
Method robscale_subset()
log2 transform and robust scale data based on subset
Usage
LFQDataTransformer$robscale_subset(
lfqsubset,
preserveMean = TRUE,
colname = "transformed_abundance"
)Method center_to_reference()
log2 transform and robust scale data based on subset
Method intensity_array()
Transforms intensities
Method intensity_matrix()
pass a function which works with matrices, e.g., vsn::justvsn
Examples
istar <- prolfqua_data('data_ionstar')$filtered()
#> Column added : nr_peptide_Id_IN_protein_Id
istar$config <- old2new(istar$config)
data <- istar$data |> dplyr::filter(protein_Id %in% sample(protein_Id, 100))
lfqdata <- LFQData$new(data, istar$config)
lfqcopy <- lfqdata$get_copy()
lfqTrans <- lfqcopy$get_Transformer()
x <- lfqTrans$intensity_array(log2)
#> Column added : log2_peptide.intensity
x$lfq$config$is_response_transformed
#> [1] TRUE
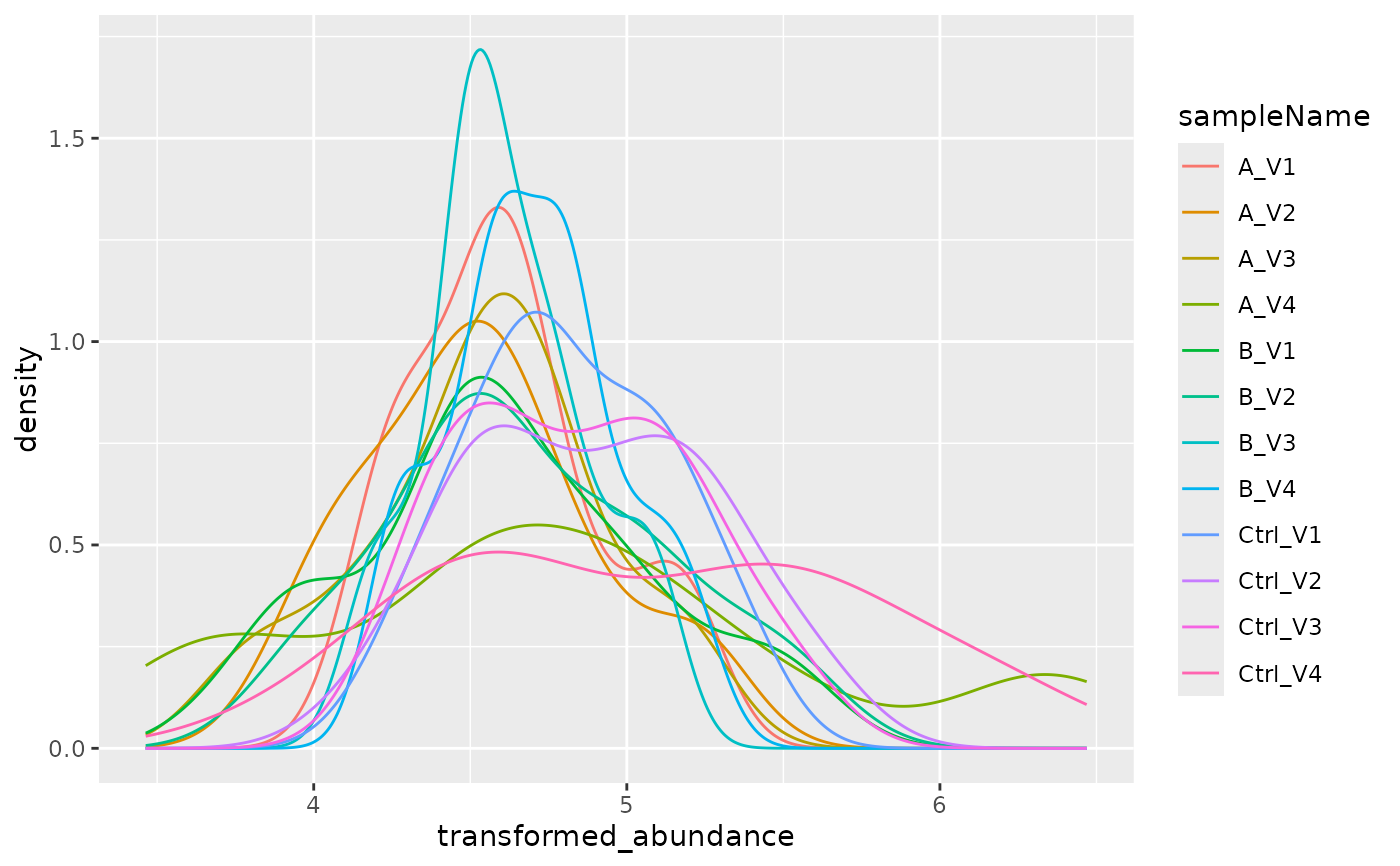
x <- x$intensity_matrix(robust_scale)
#> Warning: data already transformed. If you still want to log2 tranform, set force = TRUE
plotter <- x$lfq$get_Plotter()
plotter$intensity_distribution_density()
 # transform by asinh root and scale
lfqcopy <- lfqdata$get_copy()
lfqTrans <- lfqcopy$get_Transformer()
x <- lfqTrans$intensity_array(asinh)
#> Column added : asinh_peptide.intensity
mads1 <- mean(x$get_scales()$mads)
x <- lfqTrans$intensity_matrix(robust_scale, force = TRUE)
#> Warning: Expected 2 pieces. Additional pieces discarded in 15280 rows [1, 2, 3, 4, 5, 6,
#> 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, ...].
#> Joining with `by = join_by(protein_Id, sampleName, peptide_Id)`
mads2 <- mean(x$get_scales()$mads)
stopifnot(abs(mads1 - mads2) < 1e-8)
stopifnot(abs(mean(x$get_scales()$medians)) < 1e-8)
lfqcopy <- lfqdata$get_copy()
lfqTrans <- lfqcopy$get_Transformer()
lfqTrans$log2()
#> Column added : log2_peptide.intensity
before <- lfqTrans$get_scales()
lfqTrans$robscale()
#> data is : TRUE
#> Warning: Expected 2 pieces. Additional pieces discarded in 15280 rows [1, 2, 3, 4, 5, 6,
#> 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, ...].
#> Joining with `by = join_by(protein_Id, sampleName, peptide_Id)`
after <- lfqTrans$get_scales()
stopifnot(abs(mean(before$medians) - mean(after$medians)) < 1e-8)
stopifnot(abs(mean(before$mads) - mean(after$mads)) < 1e-8)
# normalize data using vsn
lfqcopy <- lfqdata$get_copy()
lfqTrans <- lfqcopy$get_Transformer()
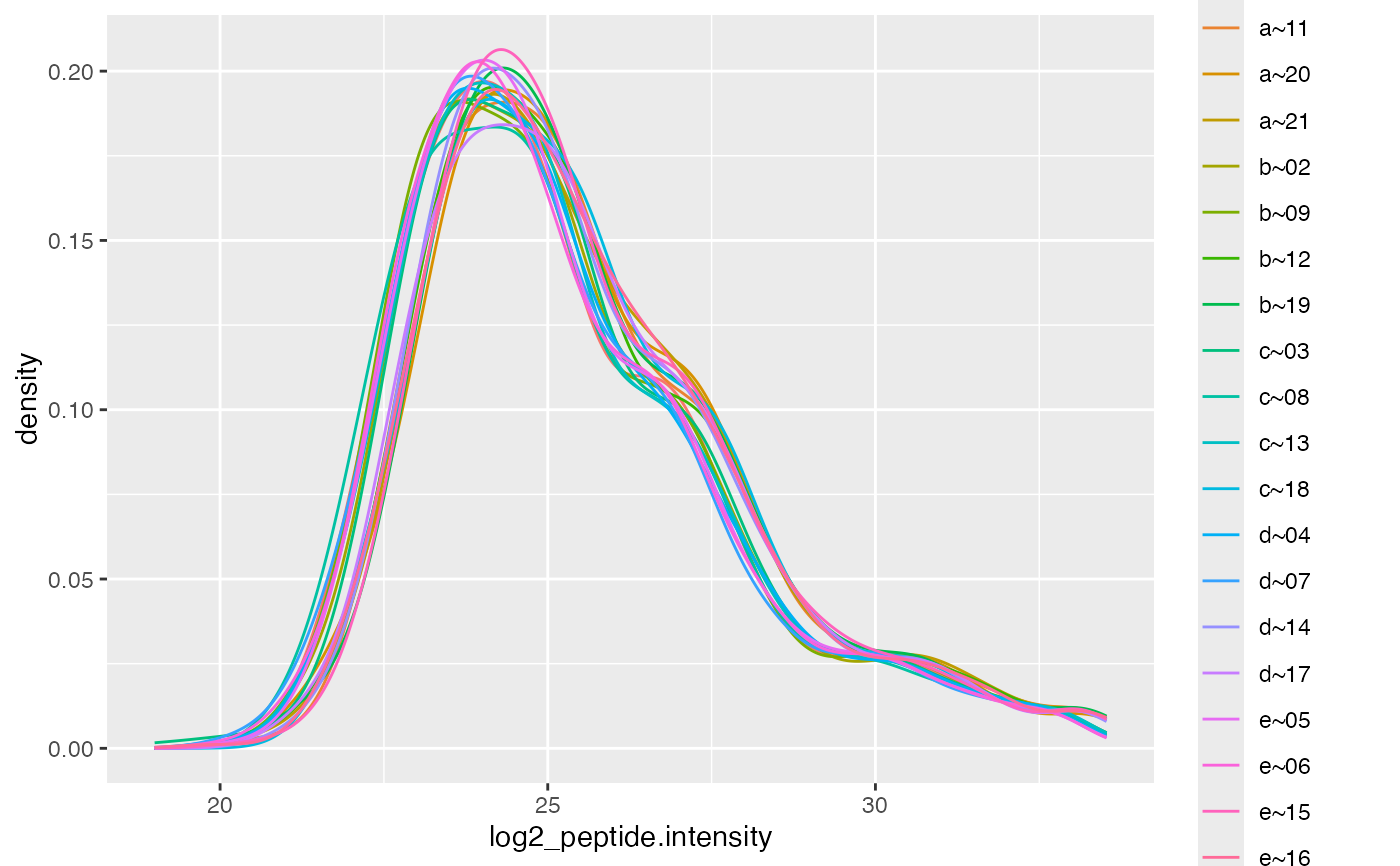
lfqTransCheck <- lfqcopy$get_Transformer()
lfqTransCheck$log2()
#> Column added : log2_peptide.intensity
lfqTransCheck$get_scales()
#> $medians
#> b~02 c~03 d~04 e~05 e~06 d~07 c~08 b~09
#> 24.86211 24.92791 24.84002 24.74686 24.74735 24.71654 24.75473 24.77979
#> a~10 a~11 b~12 c~13 d~14 e~15 e~16 d~17
#> 24.76562 25.17942 25.14736 24.87502 25.08201 25.14705 25.17601 25.19250
#> c~18 b~19 a~20 a~21
#> 25.24238 25.16130 25.18241 25.19239
#>
#> $mads
#> b~02 c~03 d~04 e~05 e~06 d~07 c~08 b~09
#> 2.250102 2.313422 2.182621 2.169717 2.165824 2.140431 2.316916 2.311747
#> a~10 a~11 b~12 c~13 d~14 e~15 e~16 d~17
#> 2.291713 2.277076 2.234688 2.221903 2.156718 2.192336 2.159619 2.295144
#> c~18 b~19 a~20 a~21
#> 2.216481 2.233216 2.232052 2.259164
#>
lfqTransCheck$lfq$get_Plotter()$intensity_distribution_density()
# transform by asinh root and scale
lfqcopy <- lfqdata$get_copy()
lfqTrans <- lfqcopy$get_Transformer()
x <- lfqTrans$intensity_array(asinh)
#> Column added : asinh_peptide.intensity
mads1 <- mean(x$get_scales()$mads)
x <- lfqTrans$intensity_matrix(robust_scale, force = TRUE)
#> Warning: Expected 2 pieces. Additional pieces discarded in 15280 rows [1, 2, 3, 4, 5, 6,
#> 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, ...].
#> Joining with `by = join_by(protein_Id, sampleName, peptide_Id)`
mads2 <- mean(x$get_scales()$mads)
stopifnot(abs(mads1 - mads2) < 1e-8)
stopifnot(abs(mean(x$get_scales()$medians)) < 1e-8)
lfqcopy <- lfqdata$get_copy()
lfqTrans <- lfqcopy$get_Transformer()
lfqTrans$log2()
#> Column added : log2_peptide.intensity
before <- lfqTrans$get_scales()
lfqTrans$robscale()
#> data is : TRUE
#> Warning: Expected 2 pieces. Additional pieces discarded in 15280 rows [1, 2, 3, 4, 5, 6,
#> 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, ...].
#> Joining with `by = join_by(protein_Id, sampleName, peptide_Id)`
after <- lfqTrans$get_scales()
stopifnot(abs(mean(before$medians) - mean(after$medians)) < 1e-8)
stopifnot(abs(mean(before$mads) - mean(after$mads)) < 1e-8)
# normalize data using vsn
lfqcopy <- lfqdata$get_copy()
lfqTrans <- lfqcopy$get_Transformer()
lfqTransCheck <- lfqcopy$get_Transformer()
lfqTransCheck$log2()
#> Column added : log2_peptide.intensity
lfqTransCheck$get_scales()
#> $medians
#> b~02 c~03 d~04 e~05 e~06 d~07 c~08 b~09
#> 24.86211 24.92791 24.84002 24.74686 24.74735 24.71654 24.75473 24.77979
#> a~10 a~11 b~12 c~13 d~14 e~15 e~16 d~17
#> 24.76562 25.17942 25.14736 24.87502 25.08201 25.14705 25.17601 25.19250
#> c~18 b~19 a~20 a~21
#> 25.24238 25.16130 25.18241 25.19239
#>
#> $mads
#> b~02 c~03 d~04 e~05 e~06 d~07 c~08 b~09
#> 2.250102 2.313422 2.182621 2.169717 2.165824 2.140431 2.316916 2.311747
#> a~10 a~11 b~12 c~13 d~14 e~15 e~16 d~17
#> 2.291713 2.277076 2.234688 2.221903 2.156718 2.192336 2.159619 2.295144
#> c~18 b~19 a~20 a~21
#> 2.216481 2.233216 2.232052 2.259164
#>
lfqTransCheck$lfq$get_Plotter()$intensity_distribution_density()
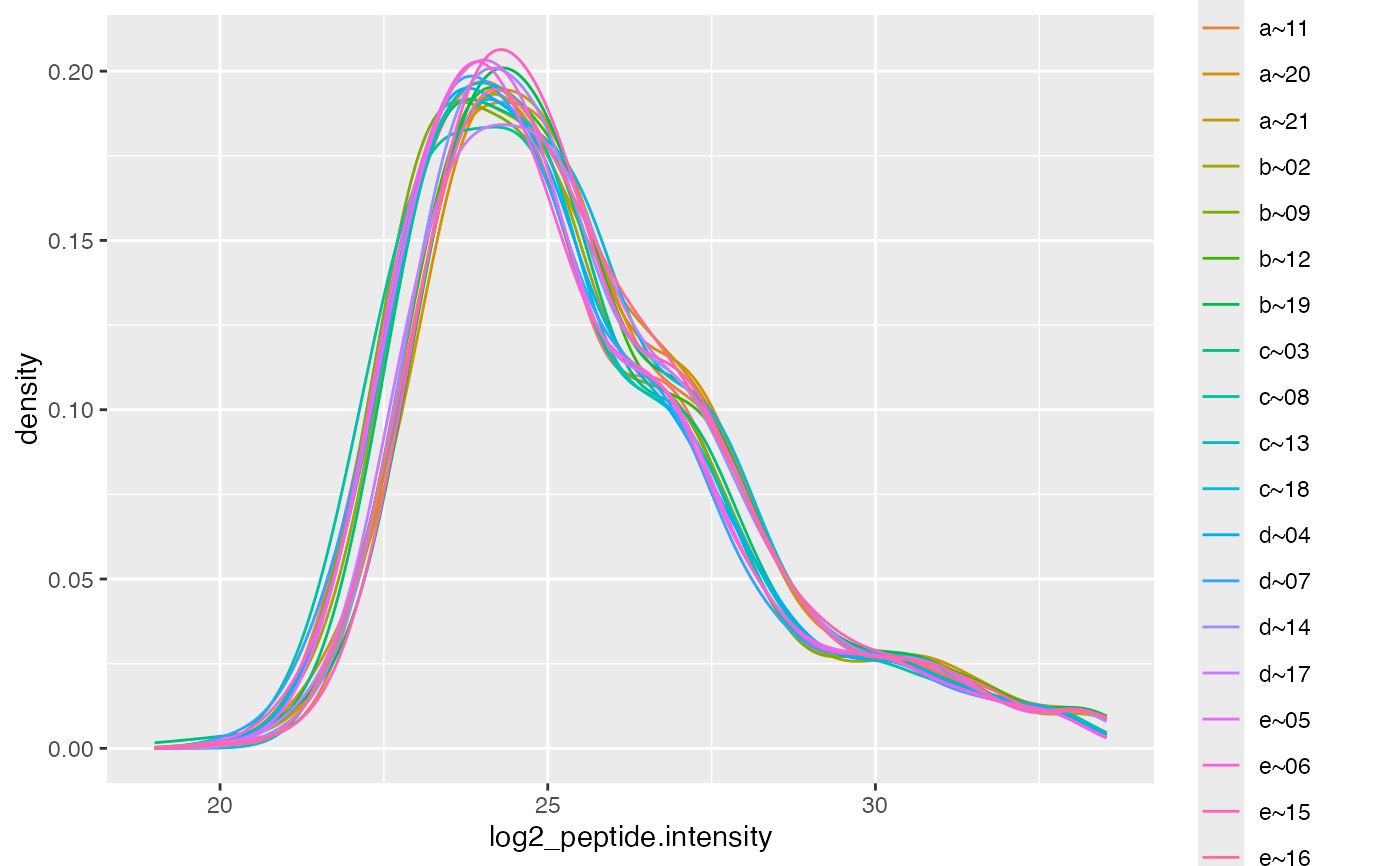 if(require("vsn")){
res <- lfqTrans$intensity_matrix( .func = vsn::justvsn)
res$lfq$get_Plotter()$intensity_distribution_density()
res$get_scales()
}
#> Loading required package: vsn
#> Warning: Expected 2 pieces. Additional pieces discarded in 15280 rows [1, 2, 3, 4, 5, 6,
#> 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, ...].
#> Joining with `by = join_by(protein_Id, sampleName, peptide_Id)`
#> $medians
#> b~02 c~03 d~04 e~05 e~06 d~07 c~08 b~09
#> 24.88535 25.00486 24.96972 24.96221 24.97661 24.95791 25.01922 24.97236
#> a~10 a~11 b~12 c~13 d~14 e~15 e~16 d~17
#> 24.94173 24.95409 24.92463 25.04857 24.92617 25.02650 25.04352 25.04521
#> c~18 b~19 a~20 a~21
#> 25.05081 24.99553 24.98337 25.03043
#>
#> $mads
#> b~02 c~03 d~04 e~05 e~06 d~07 c~08 b~09
#> 2.250102 2.313422 2.182621 2.169717 2.165824 2.140431 2.316916 2.311747
#> a~10 a~11 b~12 c~13 d~14 e~15 e~16 d~17
#> 2.291713 2.277076 2.234688 2.221903 2.156718 2.192336 2.159619 2.295144
#> c~18 b~19 a~20 a~21
#> 2.216481 2.233216 2.232052 2.259164
#>
if(require("preprocessCore")){
quant <- function(y){
ynorm <- preprocessCore::normalize.quantiles(y)
rownames(ynorm) <- rownames(y)
colnames(ynorm) <- colnames(y)
return(ynorm)
}
res <- lfqTrans$intensity_matrix( .func = quant)
res$lfq$get_Plotter()$intensity_distribution_density()
}
#> Loading required package: preprocessCore
#> Warning: data already transformed. If you still want to log2 tranform, set force = TRUE
if(require("vsn")){
res <- lfqTrans$intensity_matrix( .func = vsn::justvsn)
res$lfq$get_Plotter()$intensity_distribution_density()
res$get_scales()
}
#> Loading required package: vsn
#> Warning: Expected 2 pieces. Additional pieces discarded in 15280 rows [1, 2, 3, 4, 5, 6,
#> 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, ...].
#> Joining with `by = join_by(protein_Id, sampleName, peptide_Id)`
#> $medians
#> b~02 c~03 d~04 e~05 e~06 d~07 c~08 b~09
#> 24.88535 25.00486 24.96972 24.96221 24.97661 24.95791 25.01922 24.97236
#> a~10 a~11 b~12 c~13 d~14 e~15 e~16 d~17
#> 24.94173 24.95409 24.92463 25.04857 24.92617 25.02650 25.04352 25.04521
#> c~18 b~19 a~20 a~21
#> 25.05081 24.99553 24.98337 25.03043
#>
#> $mads
#> b~02 c~03 d~04 e~05 e~06 d~07 c~08 b~09
#> 2.250102 2.313422 2.182621 2.169717 2.165824 2.140431 2.316916 2.311747
#> a~10 a~11 b~12 c~13 d~14 e~15 e~16 d~17
#> 2.291713 2.277076 2.234688 2.221903 2.156718 2.192336 2.159619 2.295144
#> c~18 b~19 a~20 a~21
#> 2.216481 2.233216 2.232052 2.259164
#>
if(require("preprocessCore")){
quant <- function(y){
ynorm <- preprocessCore::normalize.quantiles(y)
rownames(ynorm) <- rownames(y)
colnames(ynorm) <- colnames(y)
return(ynorm)
}
res <- lfqTrans$intensity_matrix( .func = quant)
res$lfq$get_Plotter()$intensity_distribution_density()
}
#> Loading required package: preprocessCore
#> Warning: data already transformed. If you still want to log2 tranform, set force = TRUE
 #subset scaling
istar2 <- sim_lfq_data_peptide_config()
#> creating sampleName from fileName column
#> completing cases
#> completing cases done
#> setup done
lfqdata2 <- LFQData$new(istar2$data, istar2$config)
lfqdata2 <- lfqdata2$get_Transformer()$intensity_array(log2)$lfq
#> Column added : log2_abundance
head(lfqdata2$hierarchy())
#> # A tibble: 6 × 1
#> protein_Id
#> <chr>
#> 1 0EfVhX~0087
#> 2 7cbcrd~5725
#> 3 9VUkAq~4703
#> 4 BEJI92~5282
#> 5 CGzoYe~2147
#> 6 DoWup2~5896
internal <- lfqdata2$get_subset(head(lfqdata2$hierarchy()))
#> Joining with `by = join_by(protein_Id)`
internal$hierarchy()
#> # A tibble: 6 × 1
#> protein_Id
#> <chr>
#> 1 0EfVhX~0087
#> 2 7cbcrd~5725
#> 3 9VUkAq~4703
#> 4 BEJI92~5282
#> 5 CGzoYe~2147
#> 6 DoWup2~5896
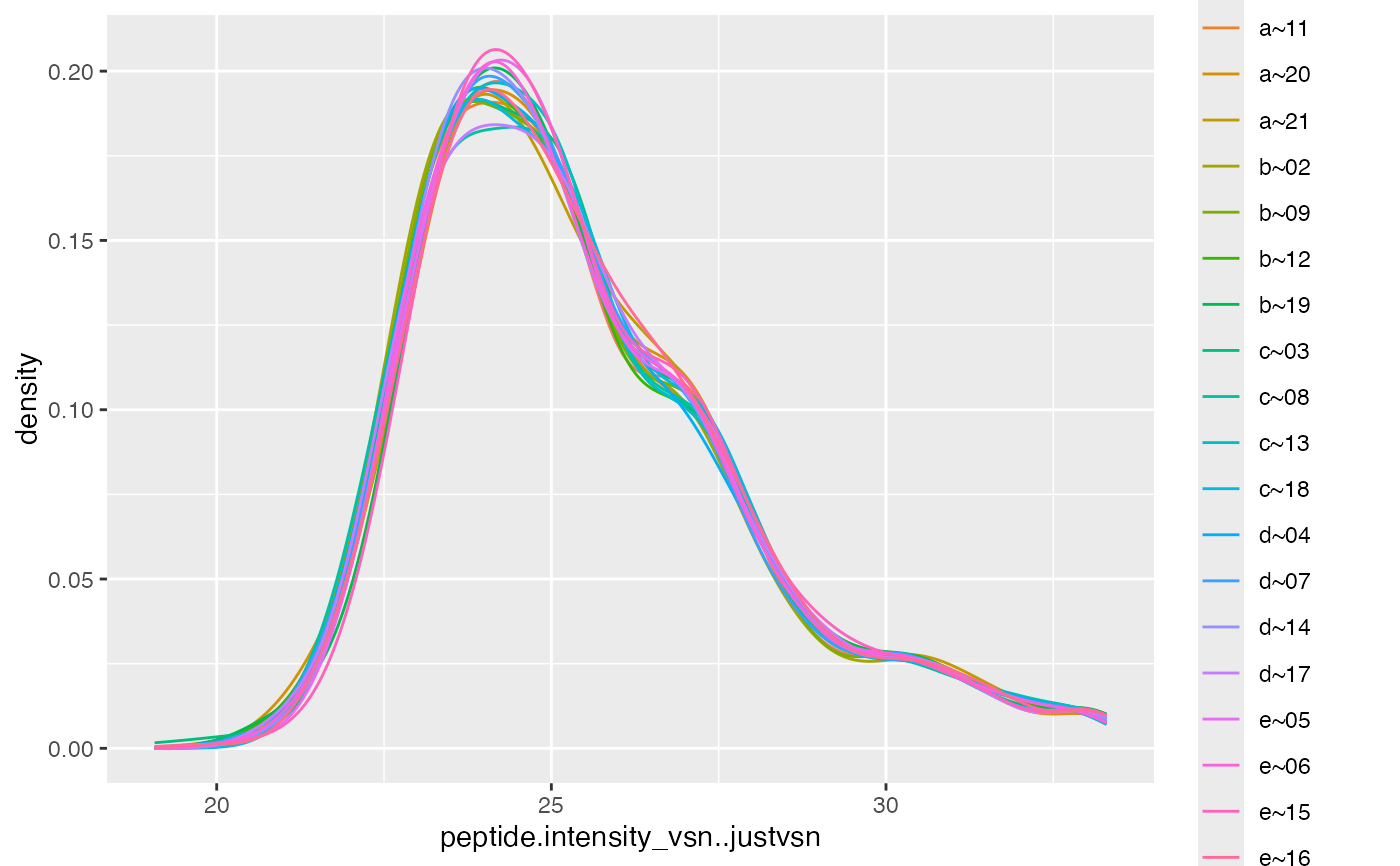
tr <- lfqdata2$get_Transformer()
tr$center_to_reference(internal)
#> data is transoformed: TRUE
pl <- tr$lfq$get_Plotter()
pl$intensity_distribution_density()
#subset scaling
istar2 <- sim_lfq_data_peptide_config()
#> creating sampleName from fileName column
#> completing cases
#> completing cases done
#> setup done
lfqdata2 <- LFQData$new(istar2$data, istar2$config)
lfqdata2 <- lfqdata2$get_Transformer()$intensity_array(log2)$lfq
#> Column added : log2_abundance
head(lfqdata2$hierarchy())
#> # A tibble: 6 × 1
#> protein_Id
#> <chr>
#> 1 0EfVhX~0087
#> 2 7cbcrd~5725
#> 3 9VUkAq~4703
#> 4 BEJI92~5282
#> 5 CGzoYe~2147
#> 6 DoWup2~5896
internal <- lfqdata2$get_subset(head(lfqdata2$hierarchy()))
#> Joining with `by = join_by(protein_Id)`
internal$hierarchy()
#> # A tibble: 6 × 1
#> protein_Id
#> <chr>
#> 1 0EfVhX~0087
#> 2 7cbcrd~5725
#> 3 9VUkAq~4703
#> 4 BEJI92~5282
#> 5 CGzoYe~2147
#> 6 DoWup2~5896
tr <- lfqdata2$get_Transformer()
tr$center_to_reference(internal)
#> data is transoformed: TRUE
pl <- tr$lfq$get_Plotter()
pl$intensity_distribution_density()
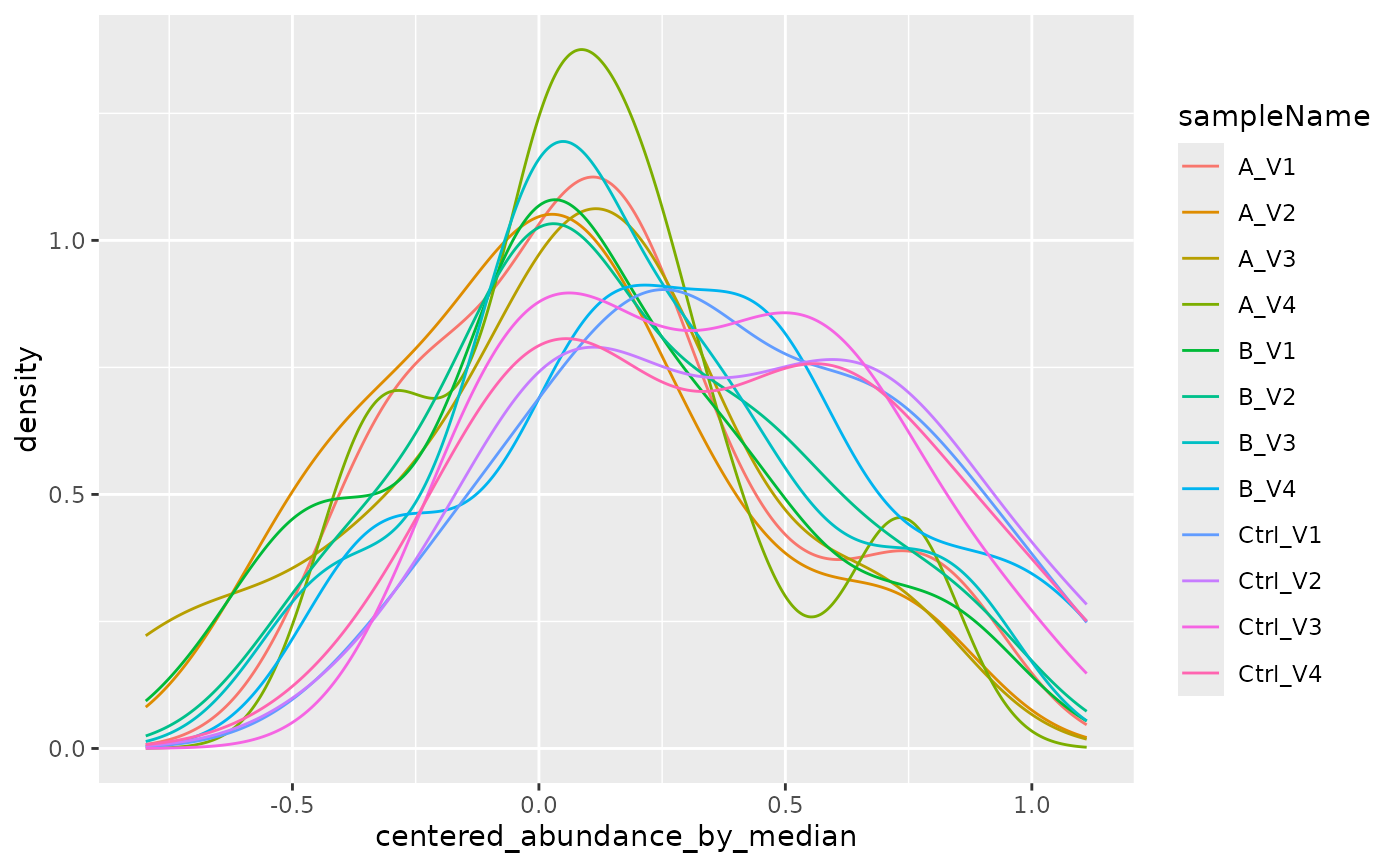 lfqdata2$get_Plotter()$intensity_distribution_density()
lfqdata2$get_Plotter()$intensity_distribution_density()
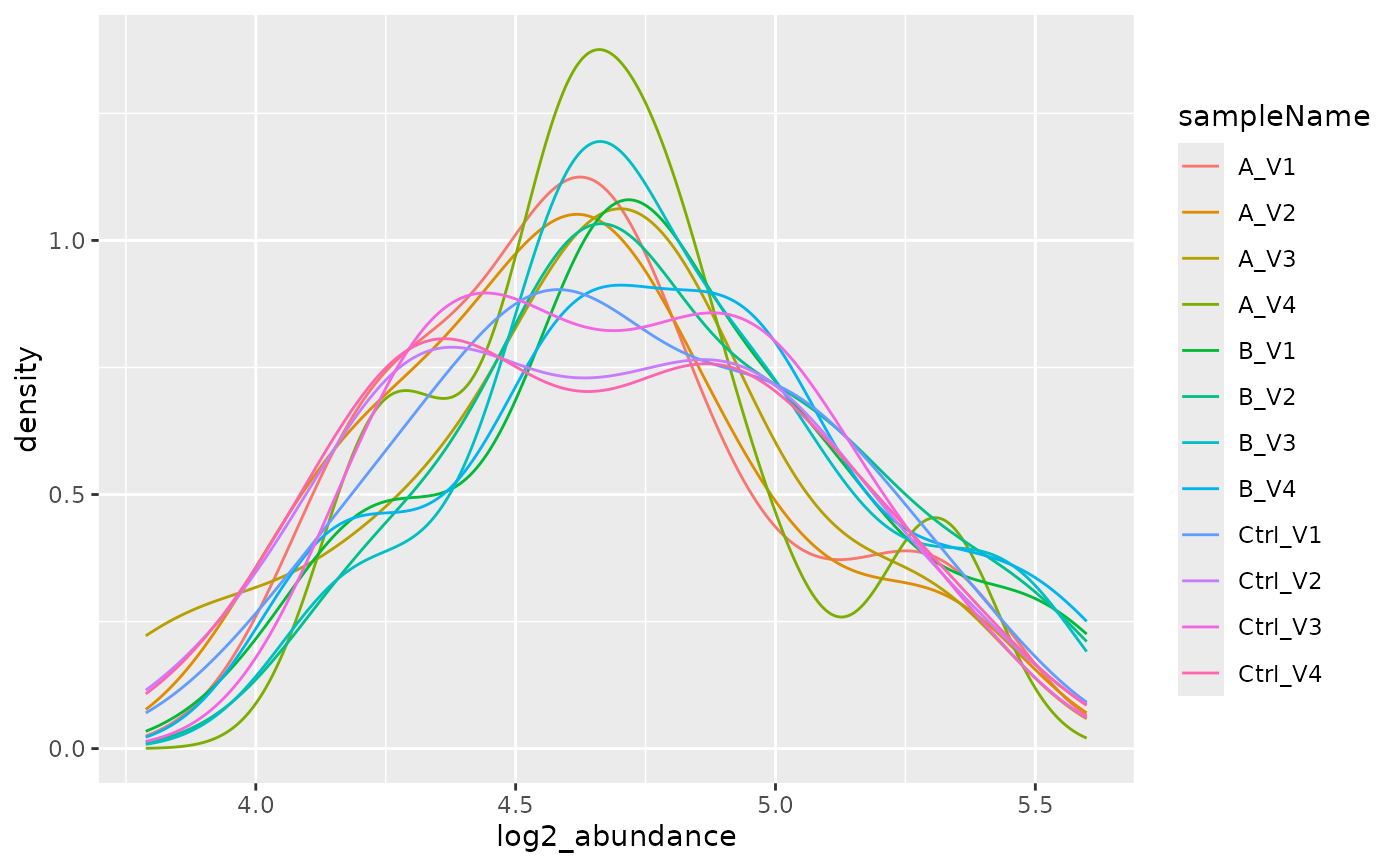 robscale <- lfqdata2$get_Transformer()$robscale_subset(internal)$lfq
#> data is : TRUE
#> Warning: Expected 2 pieces. Additional pieces discarded in 336 rows [1, 2, 3, 4, 5, 6,
#> 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, ...].
#> Joining with `by = join_by(sampleName, protein_Id, peptide_Id)`
robscale$get_Plotter()$intensity_distribution_density()
robscale <- lfqdata2$get_Transformer()$robscale_subset(internal)$lfq
#> data is : TRUE
#> Warning: Expected 2 pieces. Additional pieces discarded in 336 rows [1, 2, 3, 4, 5, 6,
#> 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, ...].
#> Joining with `by = join_by(sampleName, protein_Id, peptide_Id)`
robscale$get_Plotter()$intensity_distribution_density()