Simulate Peptide level Data
Witold E. Wolski
2026-02-25
Source:vignettes/SimulateData.Rmd
SimulateData.RmdDimulate data
knitr::opts_chunk$set(warning = FALSE, message = FALSE) For proteins: - the proteins have a FC either equal 1, 0. or -1, 10% have 1 80% have 0 and 10% have -1.
What we however are measuring are peptide spectrum matches. Let’s assume we observing peptides.
For peptides:
- The transformed protein abundances have a log normal distribution
with
meanlog = log(20), andsd = log(1.2). - The number of peptides per protein follow a geometric distribution, with
- The peptide abundances of a protein have log normal distribution
with
meanlog = log(proteinabundance)andsd = log(1.2) - The log2 intensities of a peptide within a group follow a normal distribution distribution $I_{pep} LogNormal(,) $, where is the peptide abundance and
peptideAbundances <- prolfqua::sim_lfq_data(PEPTIDE = TRUE)Analyse simulated data with prolfqua
library(prolfqua)
atable <- AnalysisConfiguration$new()
atable$fileName = "sample"
atable$factors["group_"] = "group"
atable$hierarchy[["protein_Id"]] = "proteinID"
atable$hierarchy[["peptide_Id"]] = "peptideID"
atable$set_response("abundance")
config <- AnalysisConfiguration$new(atable)
adata <- setup_analysis(peptideAbundances, config)
lfqdata <- prolfqua::LFQData$new(adata, config)
lfqdata$is_transformed(TRUE)
lfqdata$remove_small_intensities(threshold = 1)
lfqdata$filter_proteins_by_peptide_count()
lfqdata$factors()## # A tibble: 12 × 3
## sample sampleName group_
## <chr> <chr> <chr>
## 1 A_V1 A_V1 A
## 2 A_V2 A_V2 A
## 3 A_V3 A_V3 A
## 4 A_V4 A_V4 A
## 5 B_V1 B_V1 B
## 6 B_V2 B_V2 B
## 7 B_V3 B_V3 B
## 8 B_V4 B_V4 B
## 9 Ctrl_V1 Ctrl_V1 Ctrl
## 10 Ctrl_V2 Ctrl_V2 Ctrl
## 11 Ctrl_V3 Ctrl_V3 Ctrl
## 12 Ctrl_V4 Ctrl_V4 Ctrl
pl <- lfqdata$get_Plotter()
lfqdata$hierarchy_counts()## # A tibble: 1 × 3
## isotopeLabel protein_Id peptide_Id
## <chr> <int> <int>
## 1 light 16 60
lfqdata$config$hierarchy_keys_depth()## [1] "protein_Id"
pl$heatmap()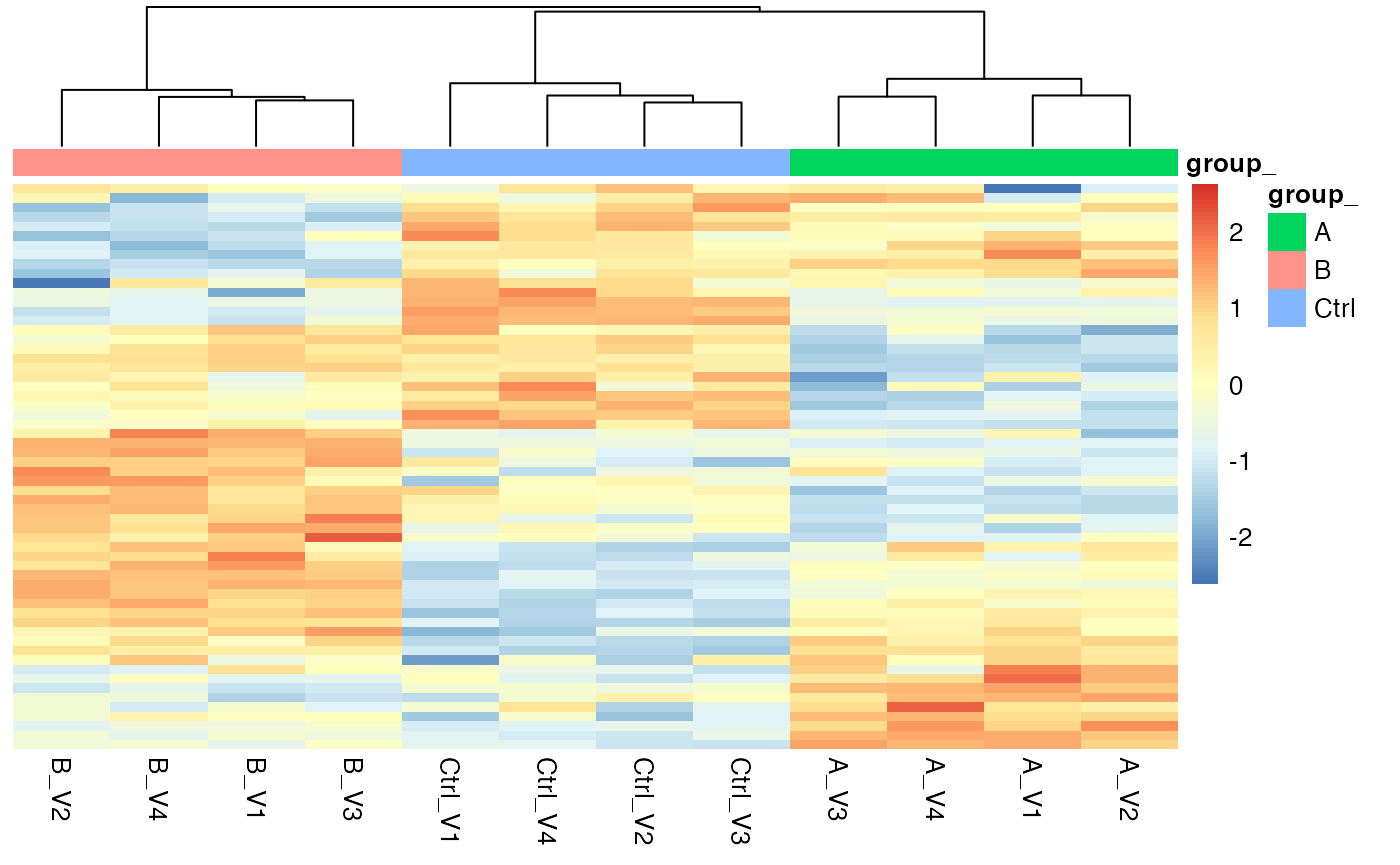
pl$intensity_distribution_density()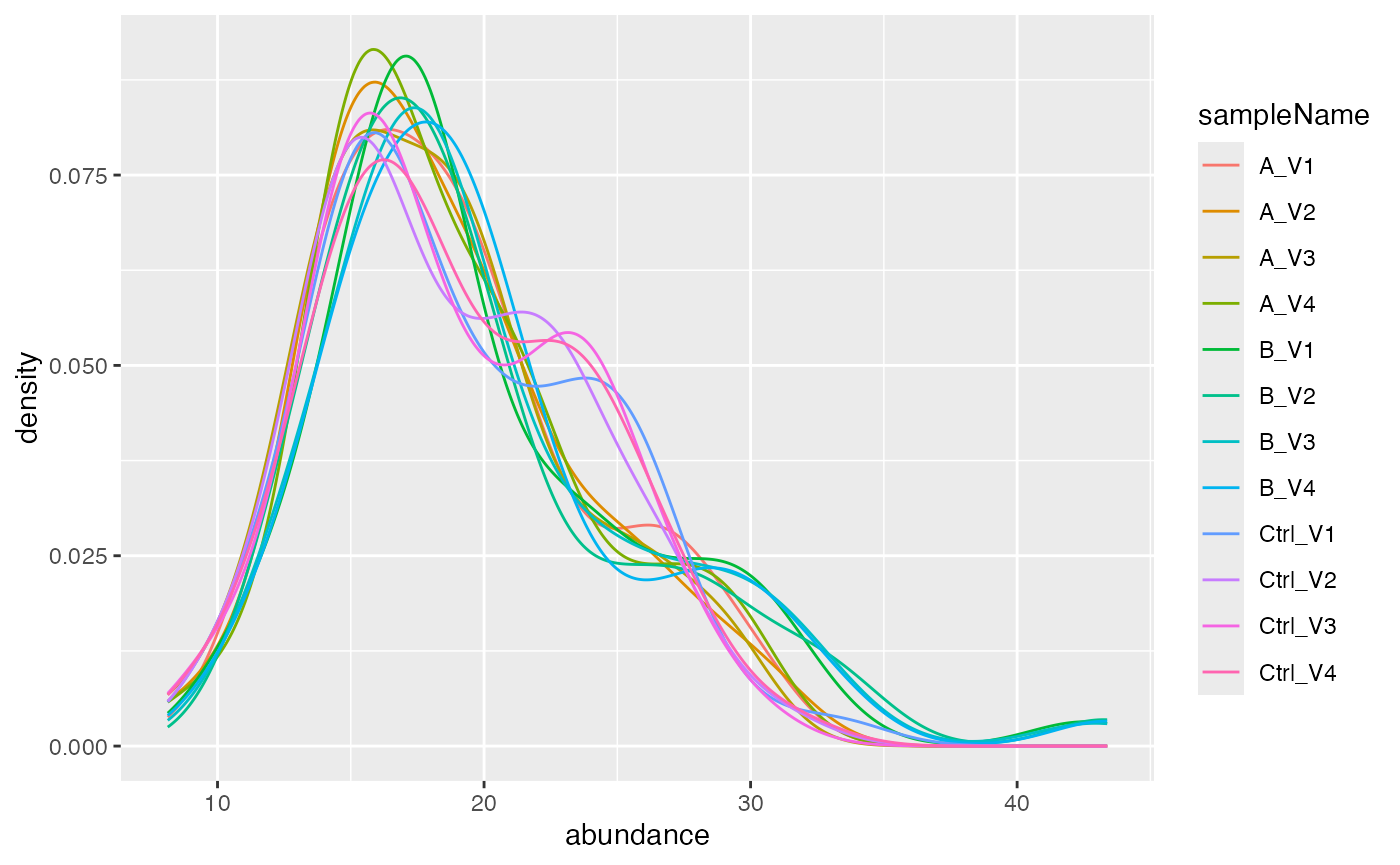
Fit peptide model
formula_Condition <- strategy_lm("abundance ~ group_")
lfqdata$config$hierarchyDepth <- 2
# specify model definition
modelName <- "Model"
contr_spec <- c("B_over_Ctrl" = "group_B - group_Ctrl",
"A_over_Ctrl" = "group_A - group_Ctrl")
lfqdata$subject_Id()## [1] "protein_Id" "peptide_Id"
mod <- prolfqua::build_model(
lfqdata,
formula_Condition)
aovtable <- mod$get_anova()
mod$anova_histogram()$plot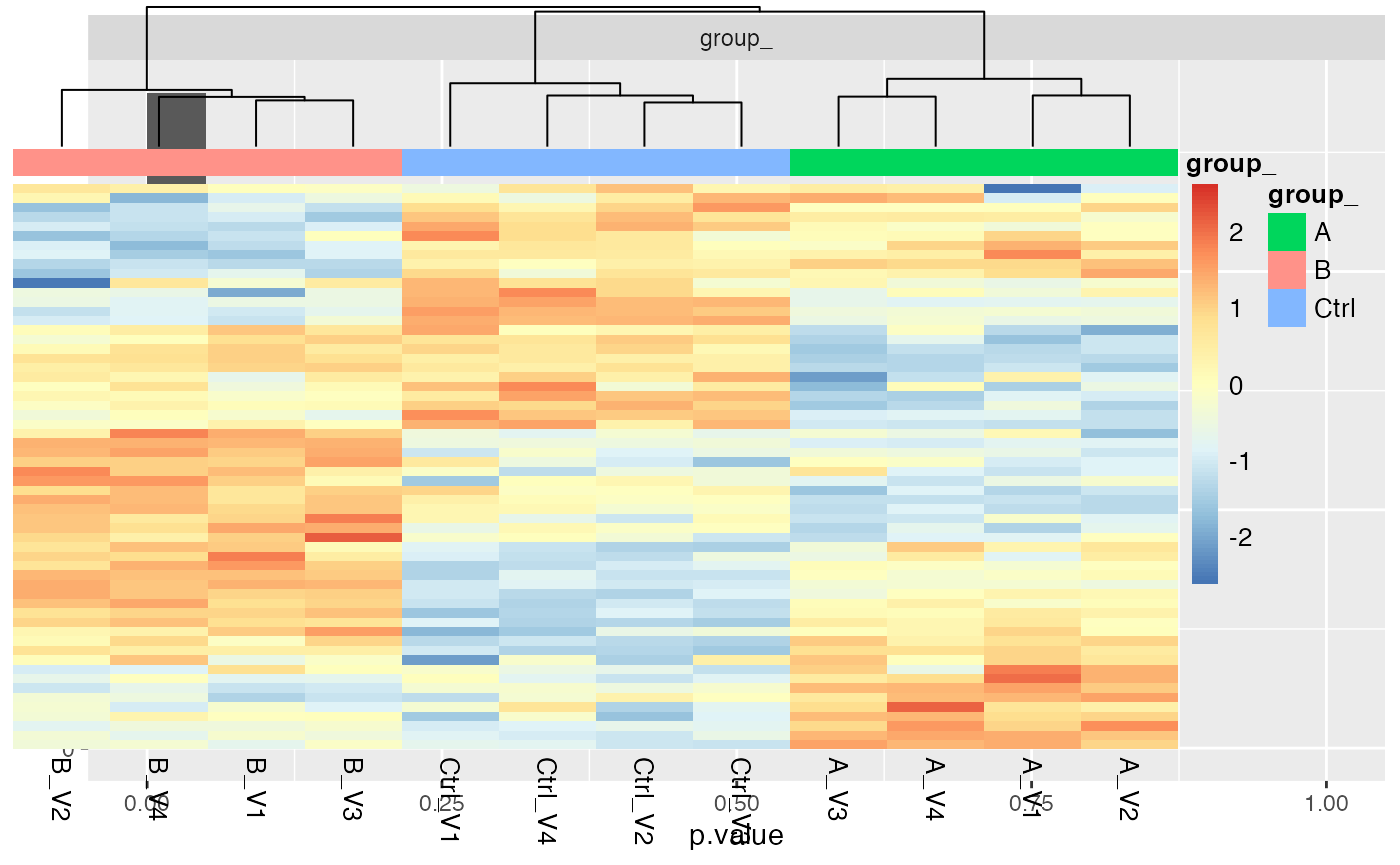
xx <- aovtable |> dplyr::filter(FDR < 0.05)
signif <- lfqdata$get_copy()
signif$data <- signif$data |> dplyr::filter(protein_Id %in% xx$protein_Id)
signif$get_Plotter()$heatmap()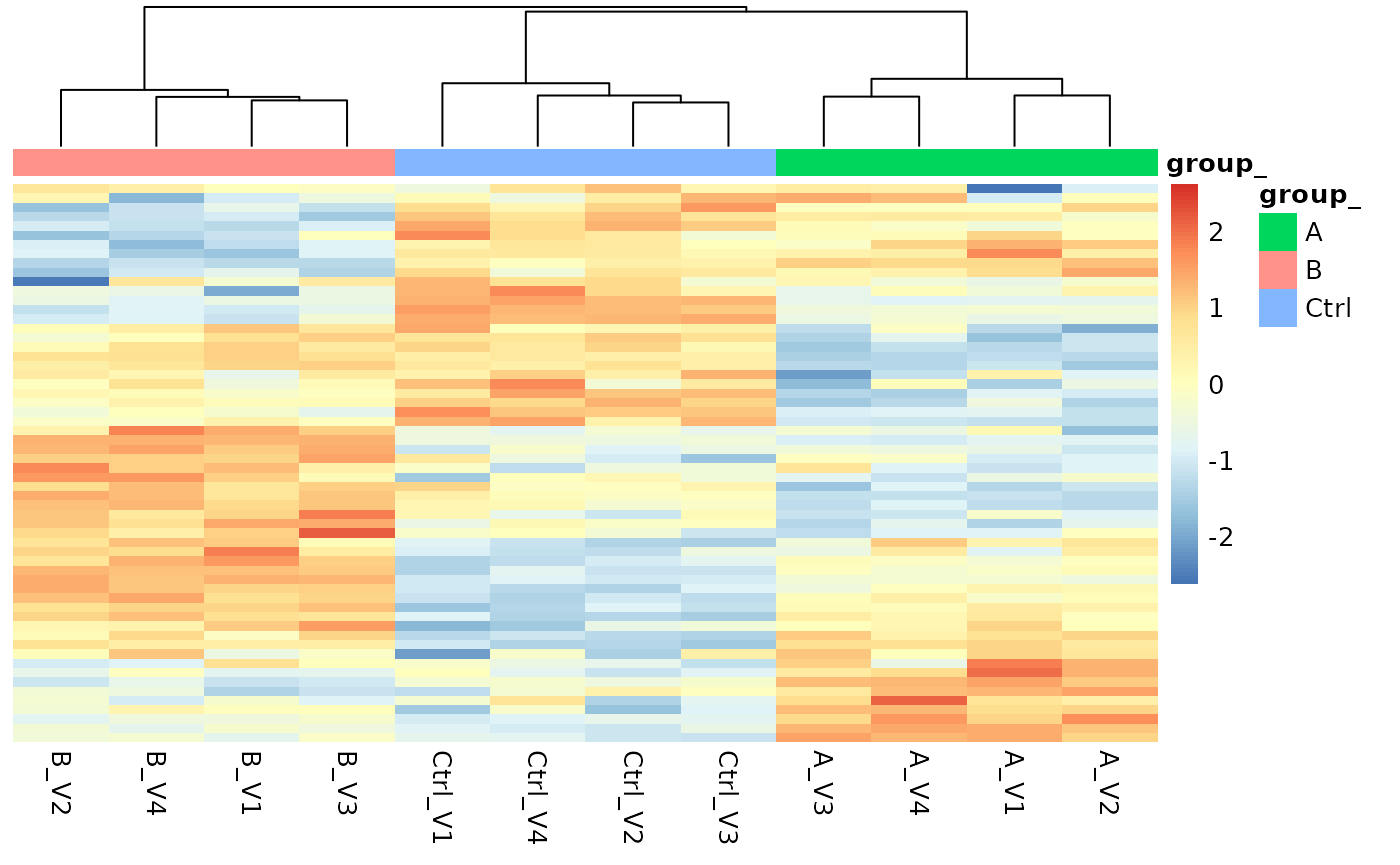
Aggregate data
lfqdata$config$hierarchyDepth <- 1
ag <- lfqdata$get_Aggregator()
ag$medpolish()
protData <- ag$lfq_agg
protData$response()## [1] "medpolish"
formula_Condition <- strategy_lm("medpolish ~ group_")
mod <- prolfqua::build_model(
protData,
formula_Condition)
contr <- prolfqua::Contrasts$new(mod, contr_spec)
v1 <- contr$get_Plotter()$volcano()
v1$FDR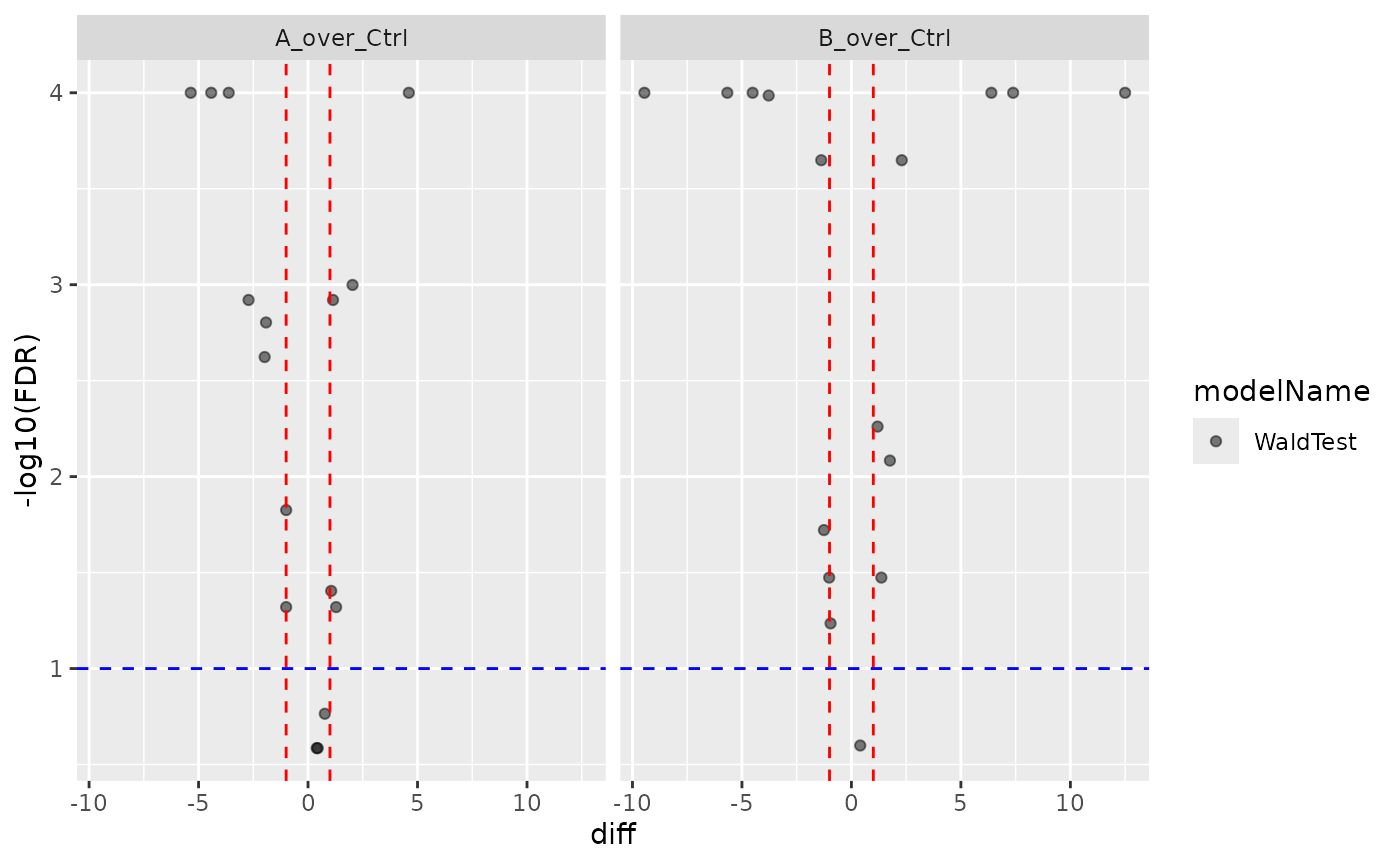
ctr <- contr$get_contrasts()