Summarize LFQData
Summarize LFQData
See also
Other LFQData:
LFQData,
LFQDataAggregator,
LFQDataPlotter,
LFQDataStats,
LFQDataToSummarizedExperiment(),
LFQDataWriter
Public fields
lfqLFQData
Methods
Method hierarchy_counts_sample()
number of elements at each level in every sample
Usage
LFQDataSummariser$hierarchy_counts_sample(
value = c("wide", "long"),
nr_children = 1
)Method plot_hierarchy_counts_sample()
barplot showing number of elements at each level in every sample
Method interaction_missing_stats()
missing per condition and protein
Method upset_interaction_missing_stats()
upset plot with missing information per protein and condition
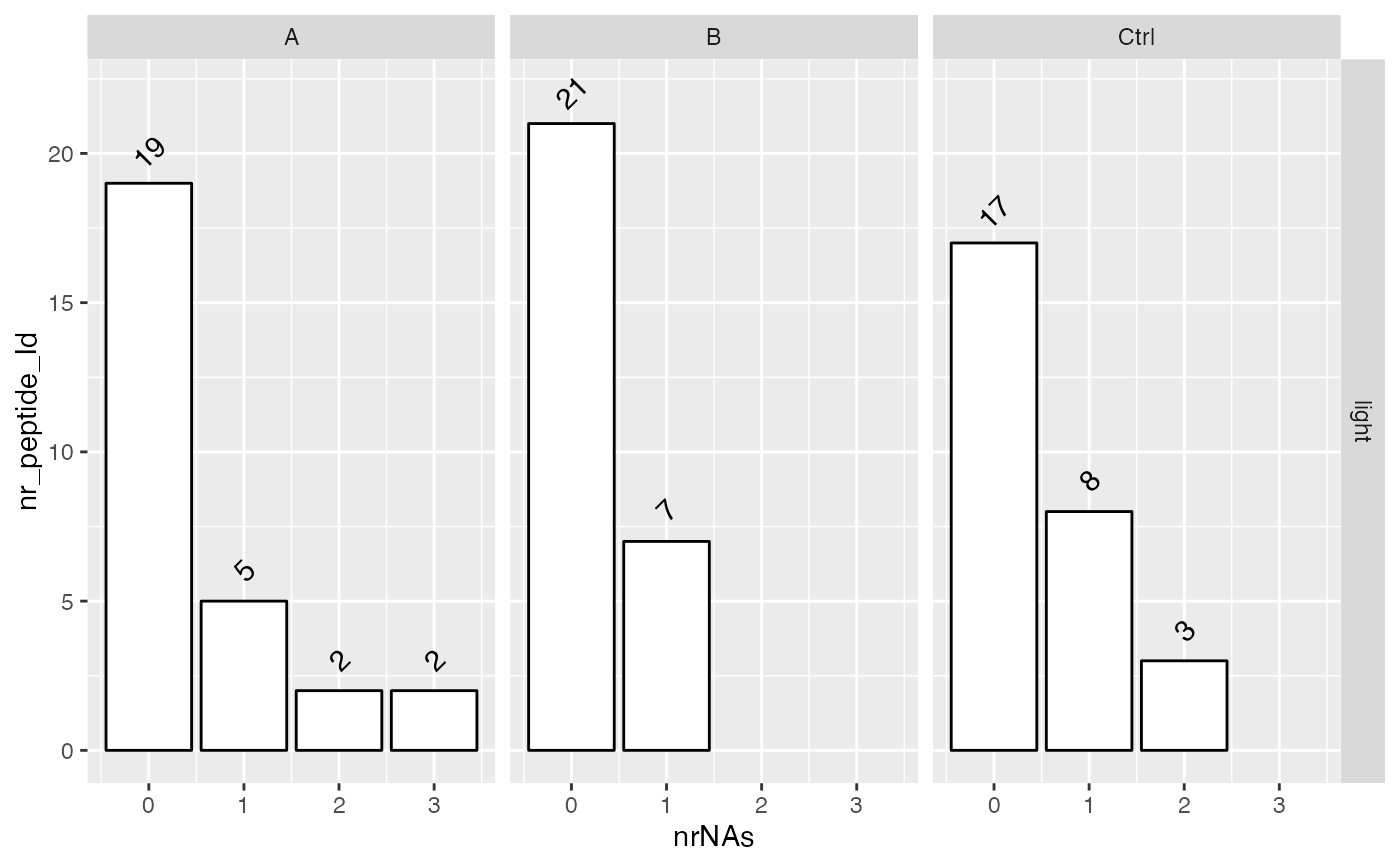
Method plot_missingness_per_group()
barplot with number of features with 1,2, etc missing in condition
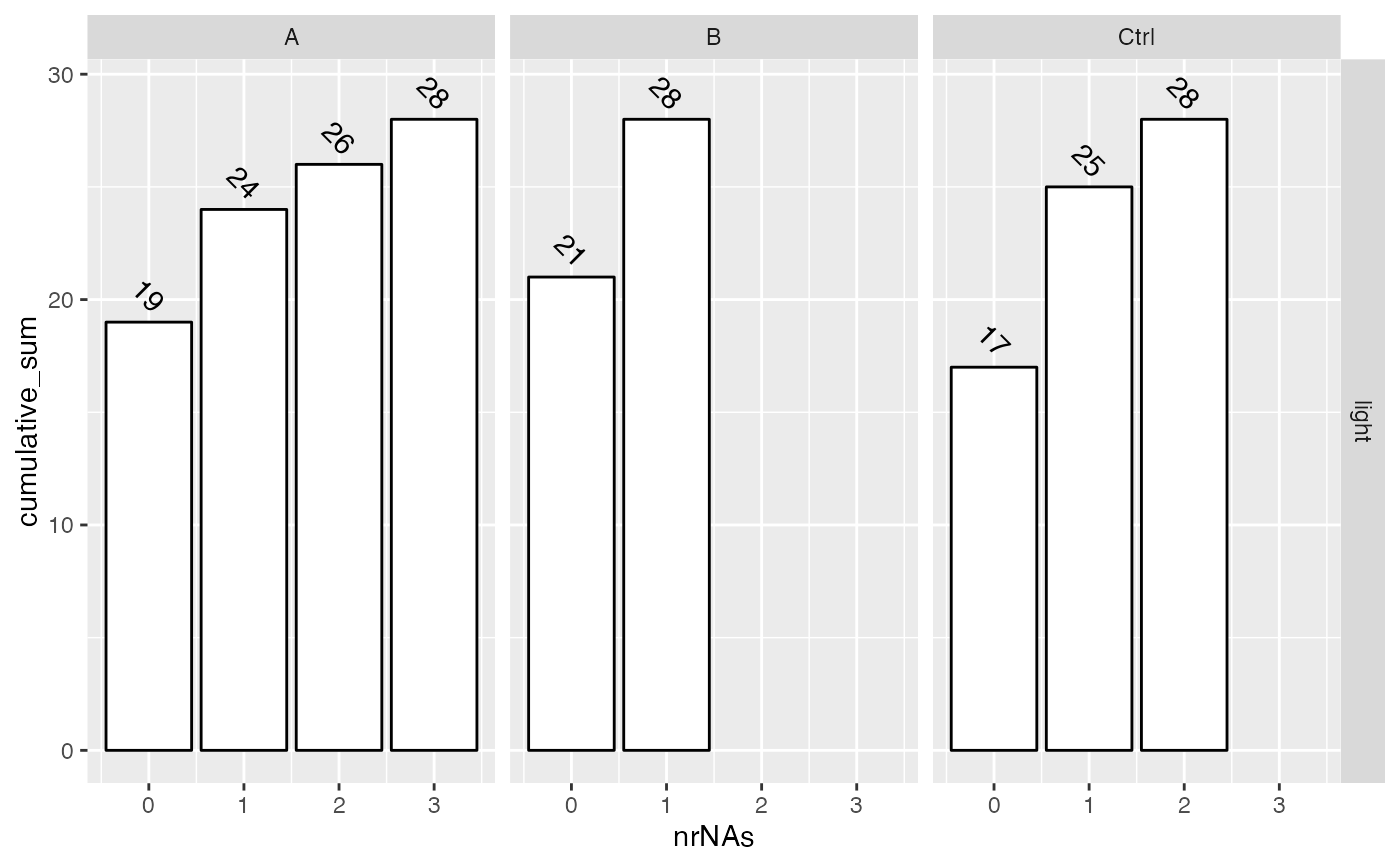
Method plot_missingness_per_group_cumsum()
barplot with cumulative sum of features with 1,2, etc missing in condition
Method percentage_abundance()
Does roll up to highest hierarchy and Computes the percent abundance of proteins overall and within each group
Examples
istar <- prolfqua::sim_lfq_data_peptide_config()
#> creating sampleName from fileName column
#> completing cases
#> completing cases done
#> setup done
data <- istar$data
lfqdata <- LFQData$new(data, istar$config)
sum <- lfqdata$get_Summariser()
sum
#> <LFQDataSummariser>
#> Public:
#> clone: function (deep = FALSE)
#> hierarchy_counts: function ()
#> hierarchy_counts_sample: function (value = c("wide", "long"), nr_children = 1)
#> initialize: function (lfqdata)
#> interaction_missing_stats: function ()
#> lfq: LFQData, R6
#> missingness_per_group: function ()
#> missingness_per_group_cumsum: function ()
#> percentage_abundance: function ()
#> plot_hierarchy_counts_sample: function (nr_children = 1)
#> plot_missingness_per_group: function ()
#> plot_missingness_per_group_cumsum: function ()
#> upset_interaction_missing_stats: function (tr = 2)
sum$hierarchy_counts()
#> # A tibble: 1 × 3
#> isotopeLabel protein_Id peptide_Id
#> <chr> <int> <int>
#> 1 light 10 28
sum$hierarchy_counts_sample("wide")
#> # A tibble: 12 × 4
#> # Groups: isotopeLabel [1]
#> isotopeLabel sampleName protein_Id peptide_Id
#> <chr> <chr> <int> <int>
#> 1 light A_V1 10 23
#> 2 light A_V2 10 26
#> 3 light A_V3 10 25
#> 4 light A_V4 10 23
#> 5 light B_V1 8 26
#> 6 light B_V2 9 26
#> 7 light B_V3 10 27
#> 8 light B_V4 10 26
#> 9 light Ctrl_V1 10 24
#> 10 light Ctrl_V2 10 25
#> 11 light Ctrl_V3 9 24
#> 12 light Ctrl_V4 10 25
sum$hierarchy_counts_sample("long")
#> # A tibble: 24 × 4
#> # Groups: isotopeLabel [1]
#> isotopeLabel sampleName key nr
#> <chr> <chr> <chr> <int>
#> 1 light A_V1 protein_Id 10
#> 2 light A_V2 protein_Id 10
#> 3 light A_V3 protein_Id 10
#> 4 light A_V4 protein_Id 10
#> 5 light B_V1 protein_Id 8
#> 6 light B_V2 protein_Id 9
#> 7 light B_V3 protein_Id 10
#> 8 light B_V4 protein_Id 10
#> 9 light Ctrl_V1 protein_Id 10
#> 10 light Ctrl_V2 protein_Id 10
#> # ℹ 14 more rows
sum$plot_hierarchy_counts_sample()
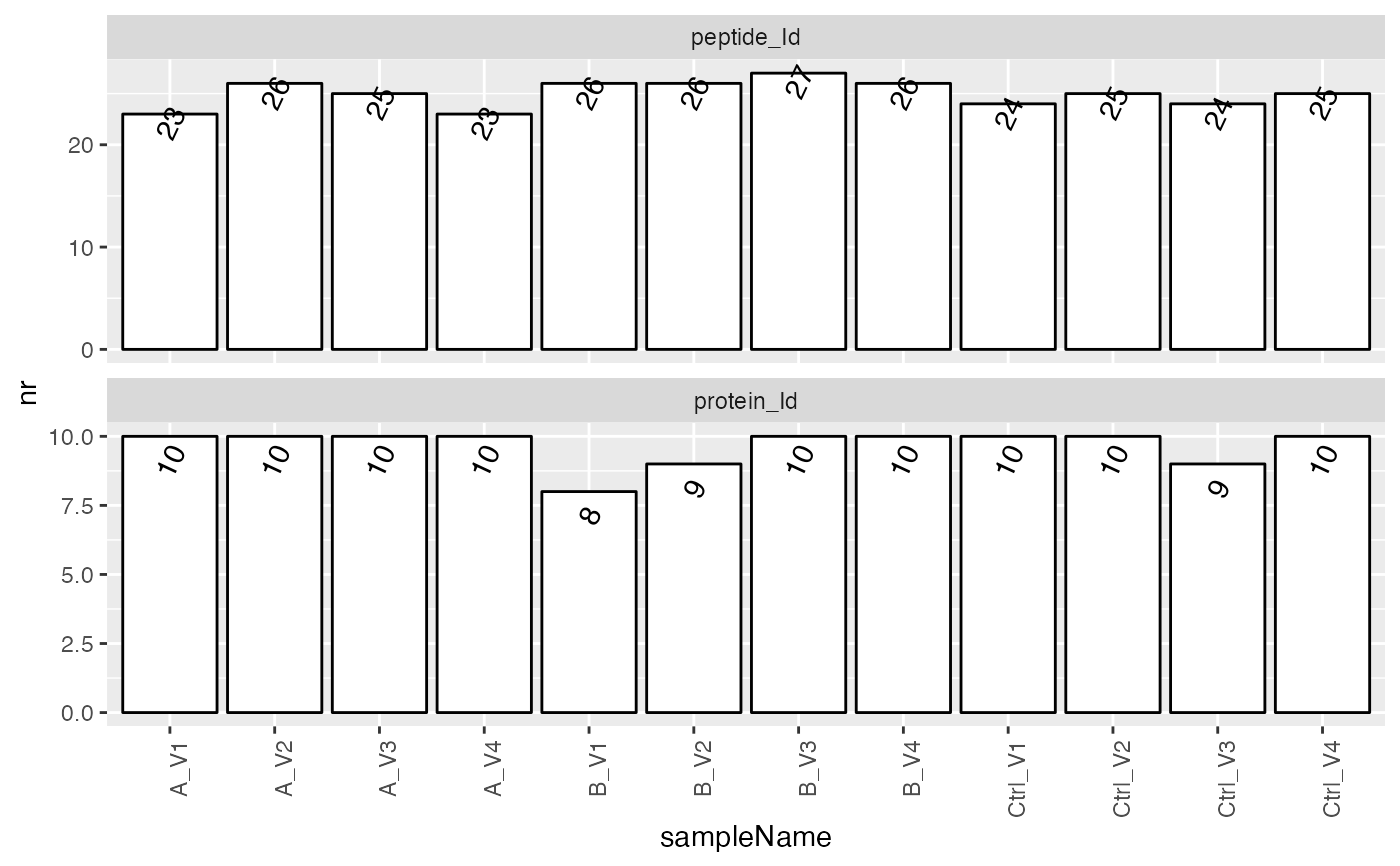 sum$plot_hierarchy_counts_sample()
sum$plot_hierarchy_counts_sample()
 tmp <- sum$interaction_missing_stats()
#> Warning: >>>> deprecated! <<<<
#>
#> use summarize_stats_factors instead.
#> completing cases
#> completing cases
sum$missingness_per_group()
#> Warning: >>>> deprecated! <<<<
#>
#> use summarize_stats_factors instead.
#> completing cases
#> # A tibble: 3 × 7
#> # Groups: isotopeLabel, group_ [3]
#> isotopeLabel group_ nrReplicates `0` `1` `2` `3`
#> <chr> <chr> <int> <int> <int> <int> <int>
#> 1 light A 4 19 5 2 2
#> 2 light B 4 21 7 NA NA
#> 3 light Ctrl 4 17 8 3 NA
sum$missingness_per_group_cumsum()
#> Warning: >>>> deprecated! <<<<
#>
#> use summarize_stats_factors instead.
#> completing cases
#> isotopeLabel ~ group_
#> # A tibble: 3 × 7
#> # Groups: isotopeLabel, group_ [3]
#> isotopeLabel group_ nrReplicates `0` `1` `2` `3`
#> <chr> <chr> <int> <int> <int> <int> <int>
#> 1 light A 4 19 24 26 28
#> 2 light B 4 21 28 NA NA
#> 3 light Ctrl 4 17 25 28 NA
sum$plot_missingness_per_group()
#> Warning: >>>> deprecated! <<<<
#>
#> use summarize_stats_factors instead.
#> completing cases
tmp <- sum$interaction_missing_stats()
#> Warning: >>>> deprecated! <<<<
#>
#> use summarize_stats_factors instead.
#> completing cases
#> completing cases
sum$missingness_per_group()
#> Warning: >>>> deprecated! <<<<
#>
#> use summarize_stats_factors instead.
#> completing cases
#> # A tibble: 3 × 7
#> # Groups: isotopeLabel, group_ [3]
#> isotopeLabel group_ nrReplicates `0` `1` `2` `3`
#> <chr> <chr> <int> <int> <int> <int> <int>
#> 1 light A 4 19 5 2 2
#> 2 light B 4 21 7 NA NA
#> 3 light Ctrl 4 17 8 3 NA
sum$missingness_per_group_cumsum()
#> Warning: >>>> deprecated! <<<<
#>
#> use summarize_stats_factors instead.
#> completing cases
#> isotopeLabel ~ group_
#> # A tibble: 3 × 7
#> # Groups: isotopeLabel, group_ [3]
#> isotopeLabel group_ nrReplicates `0` `1` `2` `3`
#> <chr> <chr> <int> <int> <int> <int> <int>
#> 1 light A 4 19 24 26 28
#> 2 light B 4 21 28 NA NA
#> 3 light Ctrl 4 17 25 28 NA
sum$plot_missingness_per_group()
#> Warning: >>>> deprecated! <<<<
#>
#> use summarize_stats_factors instead.
#> completing cases
 sum$plot_missingness_per_group_cumsum()
#> Warning: >>>> deprecated! <<<<
#>
#> use summarize_stats_factors instead.
#> completing cases
#> isotopeLabel ~ group_
sum$plot_missingness_per_group_cumsum()
#> Warning: >>>> deprecated! <<<<
#>
#> use summarize_stats_factors instead.
#> completing cases
#> isotopeLabel ~ group_
 sum$upset_interaction_missing_stats()
#> Warning: >>>> deprecated! <<<<
#>
#> use summarize_stats_factors instead.
#> completing cases
sum$upset_interaction_missing_stats()
#> Warning: >>>> deprecated! <<<<
#>
#> use summarize_stats_factors instead.
#> completing cases
 sum$percentage_abundance()
#> completing cases
#> completing cases
#> # A tibble: 112 × 16
#> # Groups: interaction [4]
#> interaction protein_Id peptide_Id group_ nrReplicates nrMeasured nrNAs
#> <chr> <chr> <chr> <chr> <int> <int> <int>
#> 1 group_A Fl4JiV~8625 fv2Ck8hz A 4 1 3
#> 2 group_A 0EfVhX~0087 dJkdz7so A 4 2 2
#> 3 group_A SGIVBl~5782 gyTlZxnW A 4 1 3
#> 4 group_A HvIpHG~9079 opjydeWJ A 4 3 1
#> 5 group_A 9VUkAq~4703 eIC06D7g A 4 4 0
#> 6 group_A BEJI92~5282 HBkZvdhT A 4 2 2
#> 7 group_A 0EfVhX~0087 ITLb4x1q A 4 3 1
#> 8 group_A HvIpHG~9079 lWjJPCzZ A 4 3 1
#> 9 group_A Fl4JiV~8625 GsUIOl6Q A 4 3 1
#> 10 group_A Fl4JiV~8625 KpyeEoiy A 4 4 0
#> # ℹ 102 more rows
#> # ℹ 9 more variables: sd <dbl>, var <dbl>, meanAbundance <dbl>,
#> # medianAbundance <dbl>, CV <dbl>, id <int>, abundance_percent <dbl>,
#> # abundance_percent_cumulative <dbl>, percent_prot <dbl>
sum$percentage_abundance()
#> completing cases
#> completing cases
#> # A tibble: 112 × 16
#> # Groups: interaction [4]
#> interaction protein_Id peptide_Id group_ nrReplicates nrMeasured nrNAs
#> <chr> <chr> <chr> <chr> <int> <int> <int>
#> 1 group_A Fl4JiV~8625 fv2Ck8hz A 4 1 3
#> 2 group_A 0EfVhX~0087 dJkdz7so A 4 2 2
#> 3 group_A SGIVBl~5782 gyTlZxnW A 4 1 3
#> 4 group_A HvIpHG~9079 opjydeWJ A 4 3 1
#> 5 group_A 9VUkAq~4703 eIC06D7g A 4 4 0
#> 6 group_A BEJI92~5282 HBkZvdhT A 4 2 2
#> 7 group_A 0EfVhX~0087 ITLb4x1q A 4 3 1
#> 8 group_A HvIpHG~9079 lWjJPCzZ A 4 3 1
#> 9 group_A Fl4JiV~8625 GsUIOl6Q A 4 3 1
#> 10 group_A Fl4JiV~8625 KpyeEoiy A 4 4 0
#> # ℹ 102 more rows
#> # ℹ 9 more variables: sd <dbl>, var <dbl>, meanAbundance <dbl>,
#> # medianAbundance <dbl>, CV <dbl>, id <int>, abundance_percent <dbl>,
#> # abundance_percent_cumulative <dbl>, percent_prot <dbl>