Aggregates e.g. protein abundances from peptide abundances
Source:R/tidyMS_aggregation.R
estimate_intensity.RdAggregates e.g. protein abundances from peptide abundances
estimate_intensity(data, config, .func)Value
returns list with data (data.frame) and config (AnalysisConfiguration)
See also
medpolish_estimate_dfconfig rlm_estimate_dfconfig
Other aggregation:
INTERNAL_FUNCTIONS_BY_FAMILY,
aggregate_intensity_topN(),
intensity_summary_by_hkeys(),
medpolish_estimate(),
medpolish_estimate_df(),
medpolish_estimate_dfconfig(),
medpolish_protein_estimates(),
plot_estimate(),
plot_hierarchies_add_quantline(),
plot_hierarchies_line(),
plot_hierarchies_line_df(),
rlm_estimate(),
rlm_estimate_dfconfig()
Examples
dd <- prolfqua::sim_lfq_data_peptide_config()
#> creating sampleName from fileName column
#> completing cases
#> completing cases done
#> setup done
config <- dd$config
data <- dd$data
data <- prolfqua::transform_work_intensity(data, config, log2)
#> Column added : log2_abundance
bbMed <- estimate_intensity(data, config, .func = medpolish_estimate_dfconfig)
#> starting aggregation
bbRob <- estimate_intensity(data, config, .func = rlm_estimate_dfconfig)
#> starting aggregation
#> Warning: 'rlm' failed to converge in 20 steps
#> Warning: 'rlm' failed to converge in 20 steps
nrow(bbMed$data)
#> [1] 116
nrow(bbRob$data)
#> [1] 116
length(bbMed$data$medpolish)
#> [1] 116
length(bbRob$data$lmrob)
#> [1] 116
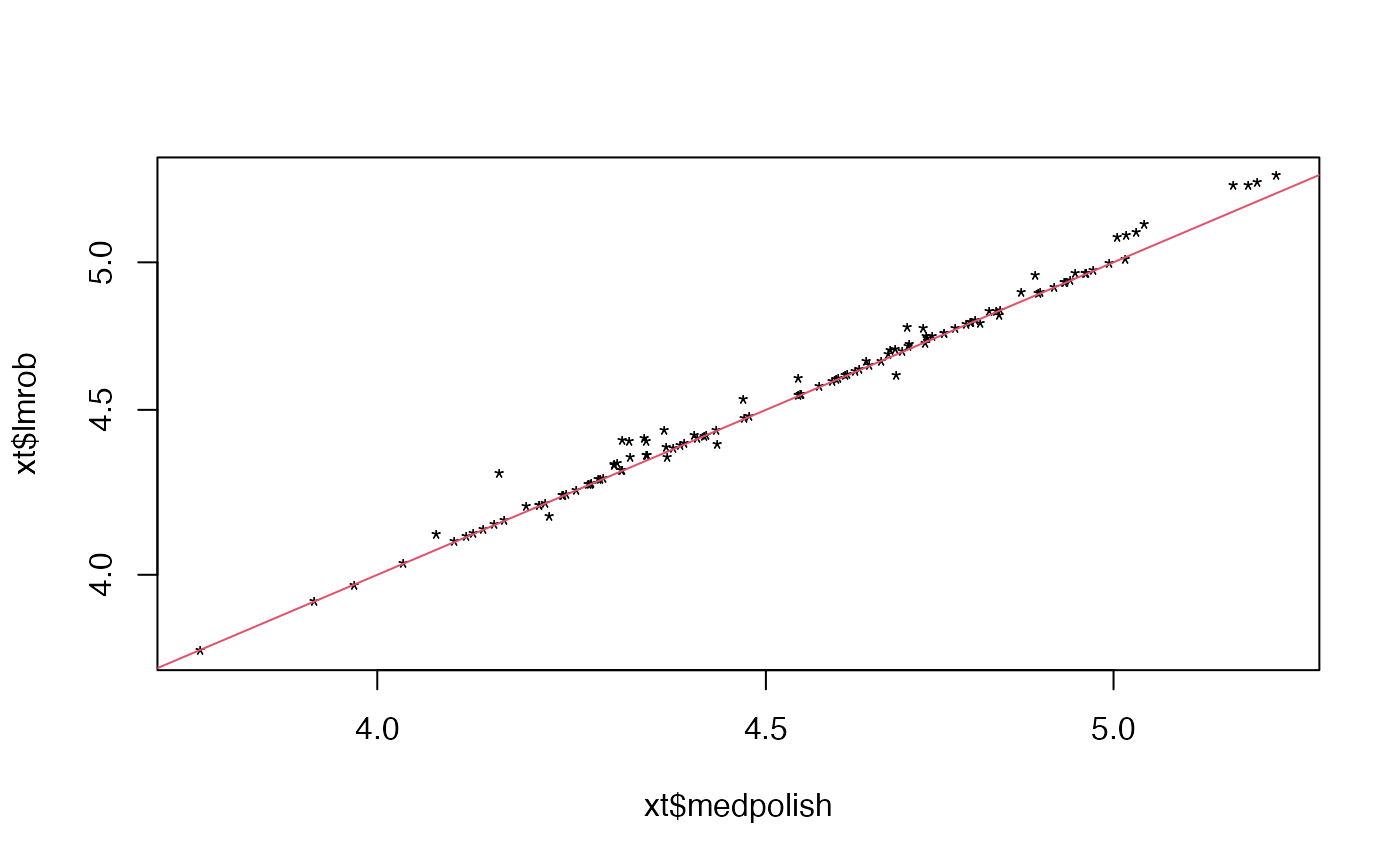
xt <- dplyr::inner_join(bbMed$data, bbRob$data)
#> Joining with `by = join_by(protein_Id, sampleName, group_, sample,
#> isotopeLabel, nr_protein_Id)`
plot(xt$medpolish, xt$lmrob, log="xy", pch="*")
abline(0,1, col=2)