plot stddev vs mean to asses stability of variance
plot_stdv_vs_mean(pdata, config, size = 2000)Arguments
See also
Examples
bb1 <- prolfqua::sim_lfq_data_peptide_config()
#> creating sampleName from fileName column
#> completing cases
#> completing cases done
#> setup done
config <- bb1$config
data <- bb1$data
res <- summarize_stats(data, config)
#> completing cases
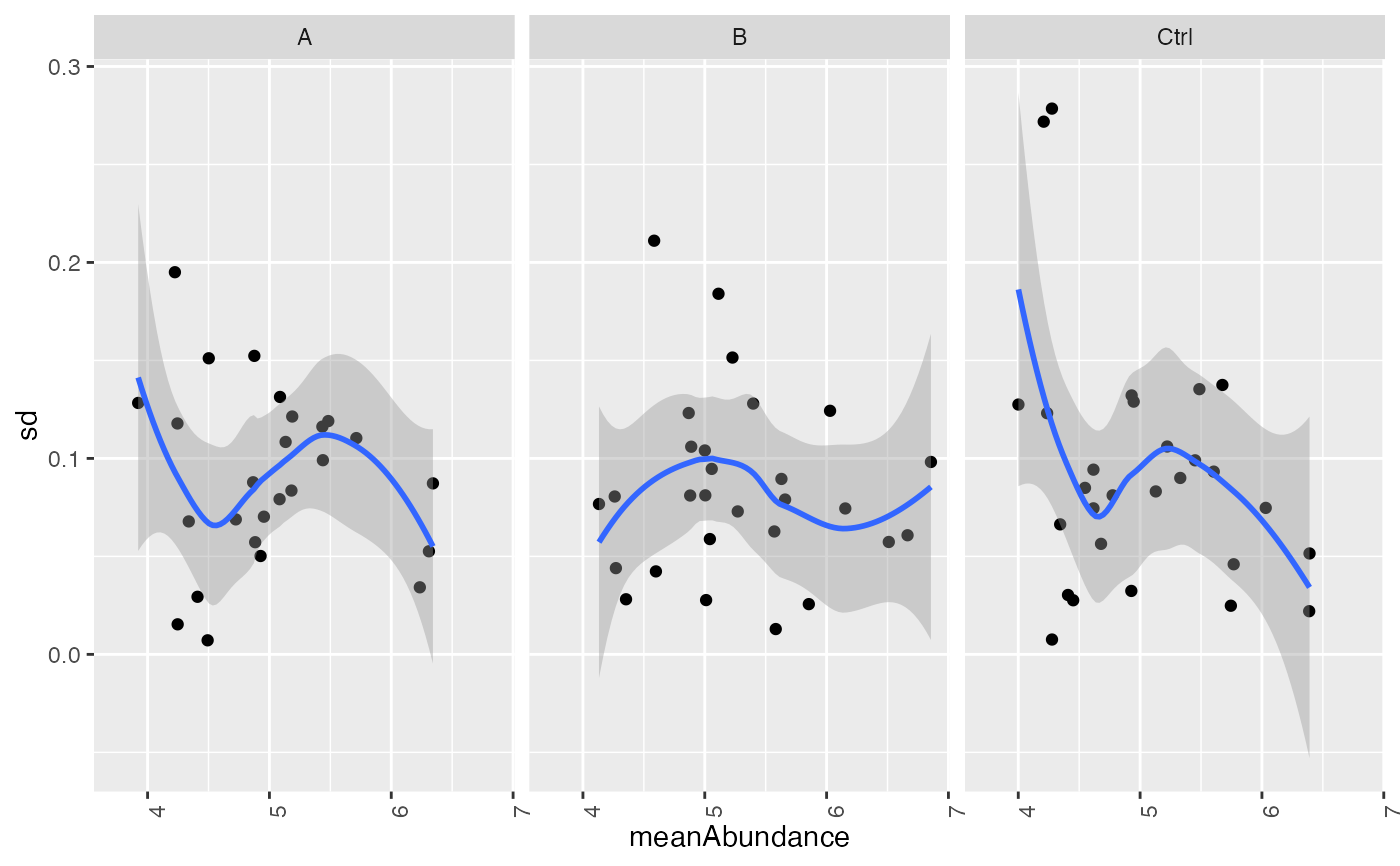
plot_stdv_vs_mean(res, config)
#> `geom_smooth()` using formula = 'y ~ x'
#> Warning: Removed 2 rows containing non-finite outside the scale range (`stat_smooth()`).
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).
 datalog2 <- transform_work_intensity(data, config, log2)
#> Column added : log2_abundance
statlog2 <- summarize_stats(datalog2, config)
#> completing cases
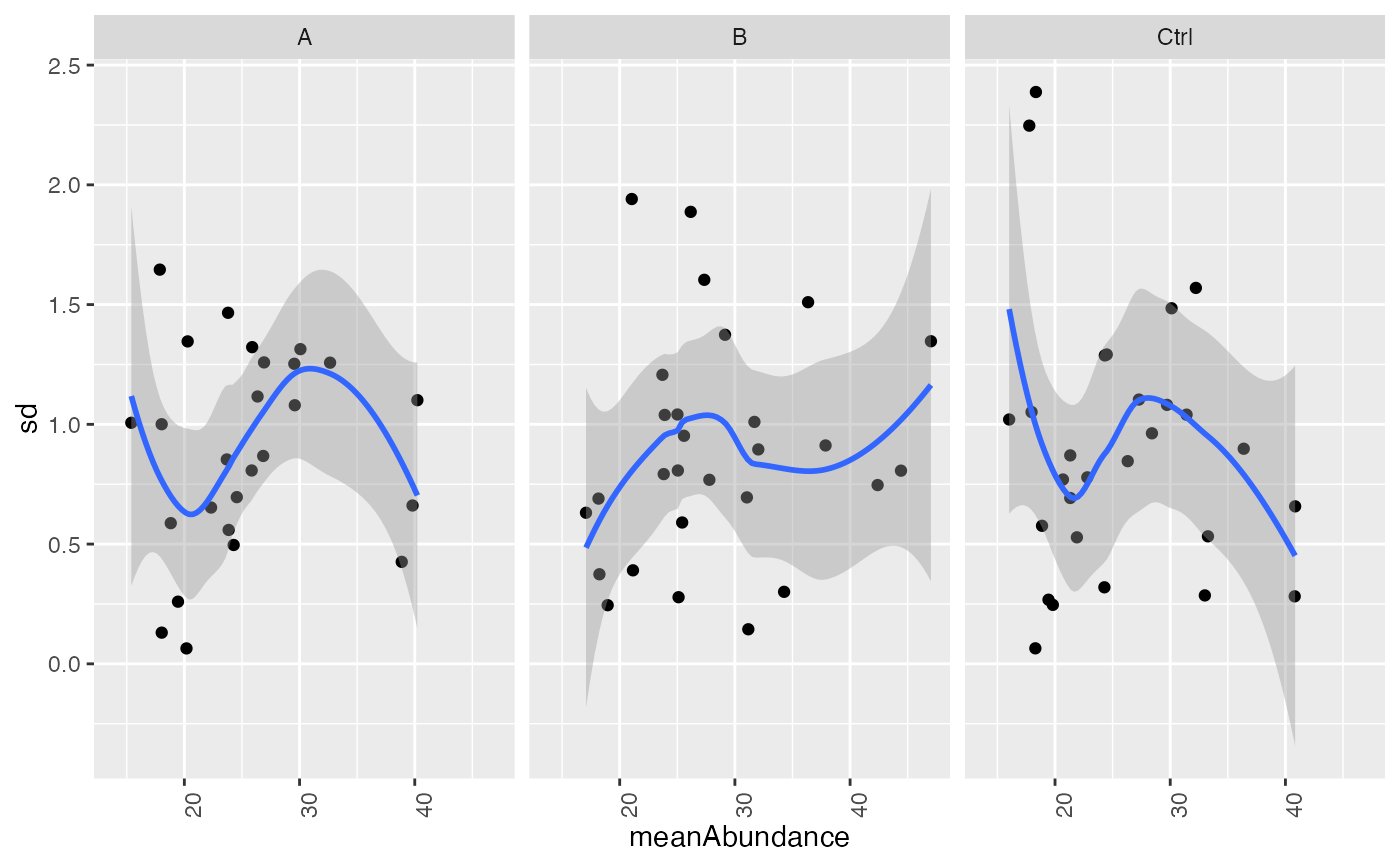
plot_stdv_vs_mean(statlog2, config)
#> `geom_smooth()` using formula = 'y ~ x'
#> Warning: Removed 2 rows containing non-finite outside the scale range (`stat_smooth()`).
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).
datalog2 <- transform_work_intensity(data, config, log2)
#> Column added : log2_abundance
statlog2 <- summarize_stats(datalog2, config)
#> completing cases
plot_stdv_vs_mean(statlog2, config)
#> `geom_smooth()` using formula = 'y ~ x'
#> Warning: Removed 2 rows containing non-finite outside the scale range (`stat_smooth()`).
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).
 config$table$get_response()
#> [1] "log2_abundance"
config$table$pop_response()
#> [1] "log2_abundance"
datasqrt <- transform_work_intensity(data, config, sqrt)
#> Column added : sqrt_abundance
ressqrt <- summarize_stats(datasqrt, config)
#> completing cases
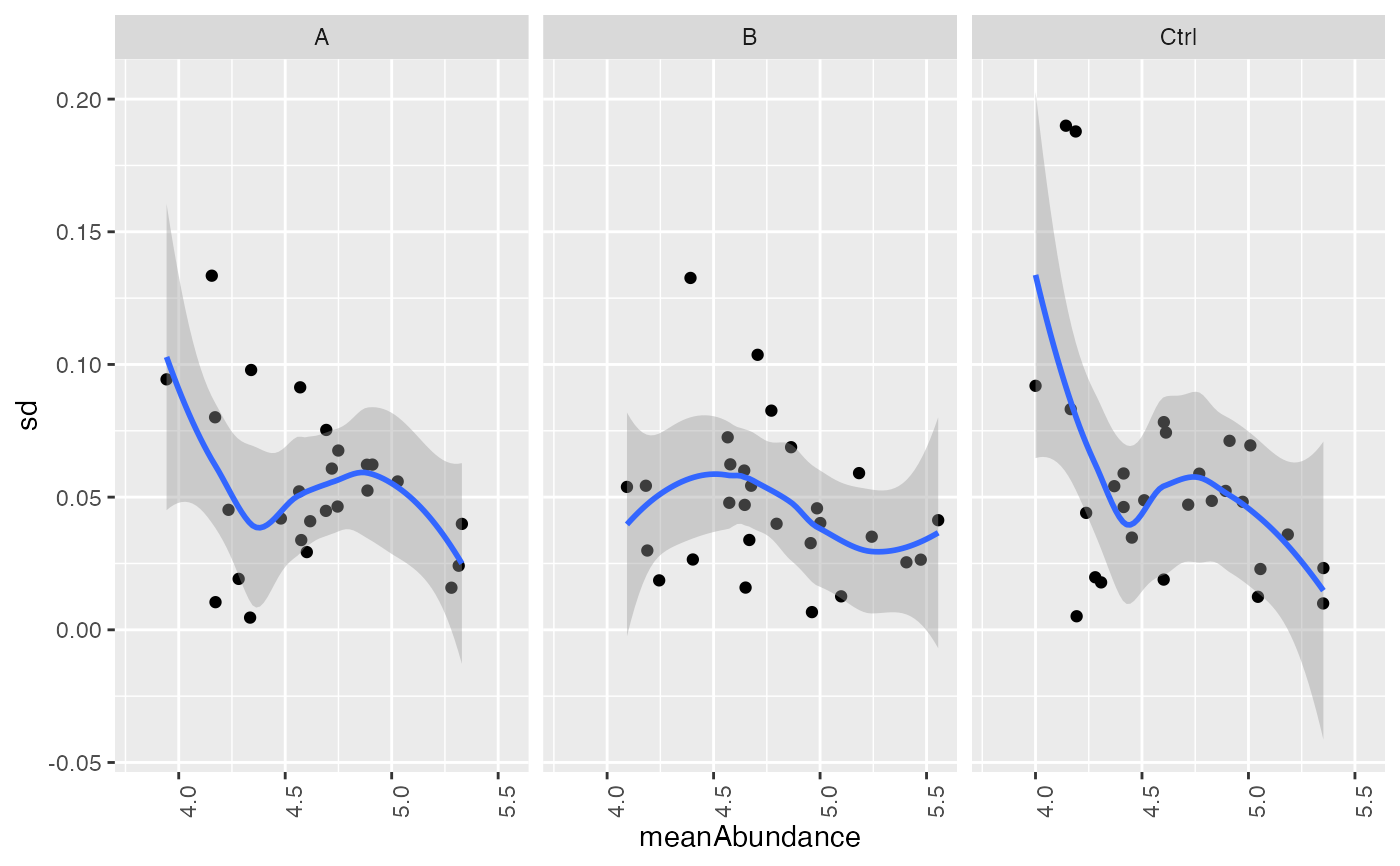
plot_stdv_vs_mean(ressqrt, config)
#> `geom_smooth()` using formula = 'y ~ x'
#> Warning: Removed 2 rows containing non-finite outside the scale range (`stat_smooth()`).
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).
config$table$get_response()
#> [1] "log2_abundance"
config$table$pop_response()
#> [1] "log2_abundance"
datasqrt <- transform_work_intensity(data, config, sqrt)
#> Column added : sqrt_abundance
ressqrt <- summarize_stats(datasqrt, config)
#> completing cases
plot_stdv_vs_mean(ressqrt, config)
#> `geom_smooth()` using formula = 'y ~ x'
#> Warning: Removed 2 rows containing non-finite outside the scale range (`stat_smooth()`).
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).